
Thus, if a node is selected on the network, then the relationship that sequences passing through that node have to other regions of diversity within the alignment can be observed through path tracing. different regions of the same sequence(s).
#Highlight sequence clc sequence viewer windows#
Edges are placed between nodes of neighbouring windows where they share sequence identification(s), i.e. Within the network, nodes are based on the clustering of sequence fragments that span windows placed consecutively along the alignment. We present CView, a visualizer that expands on the per-site representation of residues through the incorporation of a dynamic network that is based on the summarization of diversity present across different regions of the alignment. A basic alignment viewer can have potential to increase initial insight through visual enhancement, whilst not delving into the realms of complex sequence analysis. However, the initial insight gained is limited, something that is especially true when viewing alignments consisting of many sequences representing differing factors such as location, date and subtype. The persistence of this tendency to the recent tools designed for viewing mapped read data indicates that such a perspective not only provides a reliable visualization of per-site alterations, but also offers implicit reassurance to the end-user in relation to data accessibility.
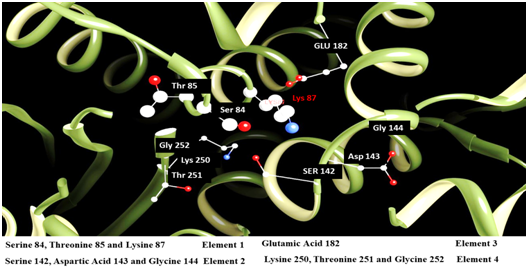
To date basic visualization of sequence alignments have largely focused on displaying per-site columns of nucleotide, or amino acid, residues along with associated frequency summarizations.


 0 kommentar(er)
0 kommentar(er)
